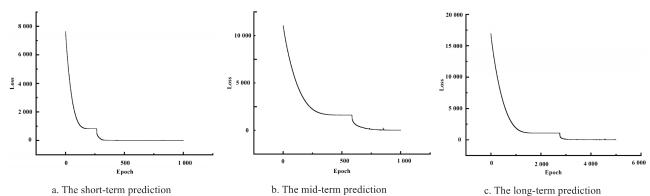
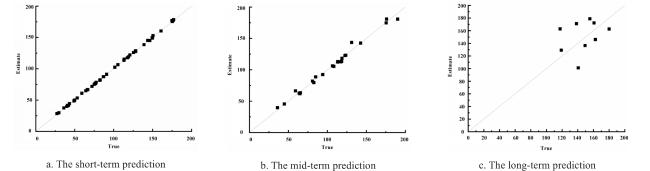
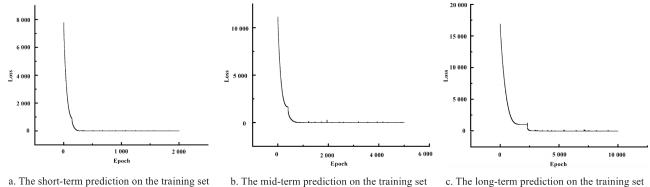
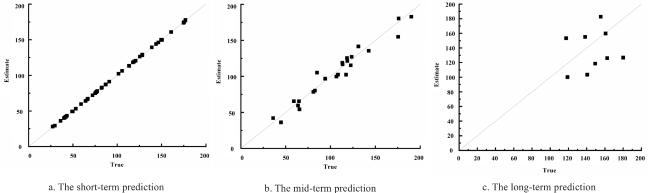
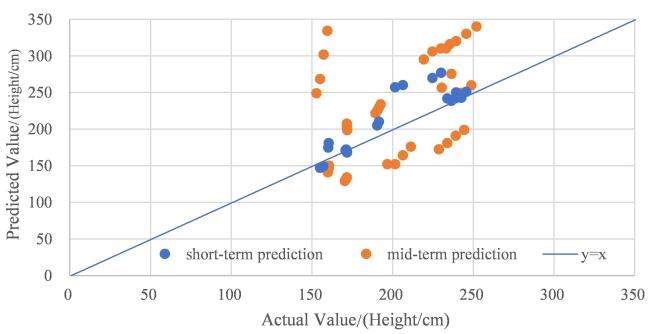
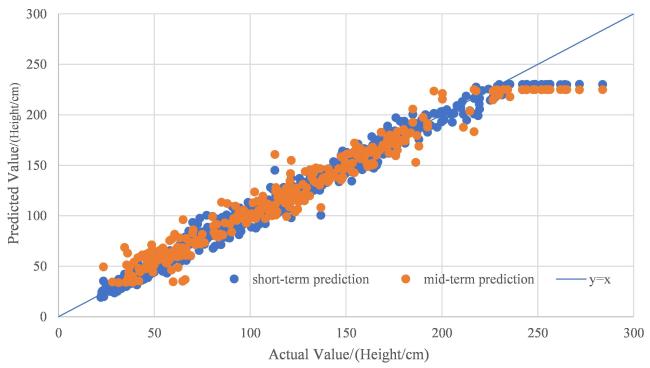
The integration of machine learning with crop models has experienced rapid development, falling within the realm of empirical models and showcasing strong practicality. In the early stages of this integration, research predominantly focused on the application of BP neural networks. Han et al.
[5] successfully developed a principal component analysis-backpropagation neural network (PCA-BPNN) network to predict fruit diameter based on air temperature, air humidity, soil moisture content, and leaf temperature, achieving a maximum error of only 0.075 mm. Focusing on flowering tomatoes, Zhang et al.
[6] established a prediction model for environmental factors and photosynthetic rate using a BP network. The models fitted under various conditions achieved a high accuracy, with an
R 2 value of 0.96 between the highest predicted and actual values. Additionally, BP network-based models have been successfully applied in predicting winter wheat water consumption
[7], conducting comprehensive evaluation of soil nutrient models
[8], and modeling CO
2 concentration models for the growth of shiitake mushrooms
[9]. All these applications have yielded promising results, highlighting the potential of machine learning in agricultural modeling. Deep learning has revolutionized the integration of machine learning with crop models, particularly through advanced image processing and the incorporation of environmental data. By leveraging deep networks and convolutional methods, deep learning achieves high accuracy in crop growth analysis. For instance, Chen et al.
[10] combined structural and near-infrared image features, identifying random forest as the optimal approach for biomass prediction. Yang et al.
[11] used convolutional neural network (CNN) with random forests and depth maps, thereby reducing the normalized mean square error (NMSE) in lettuce growth predictions. Chandel et al.
[12] demonstrated that GoogleNet excelled in classifying water stress, while Yalcin
[13] used activation maps derived from sunflower images for yield estimation. Beyond image analysis, deep learning also excels when working solely with environmental data. Bali et al.
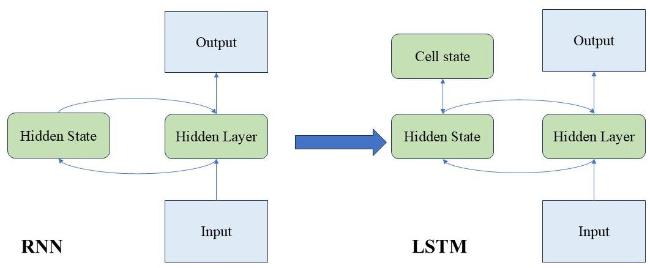
[14] showed that long short-term memory (LSTM) models outperformed other methods in predicting wheat yield using historical climate data. Nigam et al.
[15] assessed various deep learning models for yield prediction based on temperature and rainfall. Elavarasan and Vincentt
[16] achieved 93.7% accuracy in crop yield prediction with a Q-network model. In tomato yield prediction, De Alwis et al.
[17] proposed a domain adaptation long short-term memory (DA-LSTM) model that outperformed traditional LSTM, XGBoost Regression (XGBR), and support vector regression (SVR) models. Zhou et al.
[18] integrated the TOMSIM and GreenLight models with deep neural network(DNN) to improve predictions of greenhouse climate-tomato production. Statistics revealed a 30-fold increase in the use of deep learning for crop yield prediction from 2016 to 2020
[19], often surpassing the performance of traditional models
[20]. However, deep learning's reliance on specific datasets limits its transferability between different crops or regions
[21, 22], necessitating extensive localized data to address regional variation and ensure model adaptability.